Revista de la Facultad de Ciencias
Agrarias. Universidad Nacional de Cuyo. Tomo 56(2). ISSN (en línea) 1853-8665.
Año 2024.
Original article
First
report of the causal agent of vine crown gall in Mendoza, Argentina
Primer
reporte del agente causal de la agalla de corona de la vid en Mendoza,
Argentina
Mariano Emanuel
Diaz1,
1INTA EEA Mendoza. Laboratorio de Fitopatología. San Martín 3853.
Luján de Cuyo. C. P. 5507.
2Universidad Nacional de Cuyo. Facultad de Ciencias Agrarias.
Cátedra de Industrias Agrarias FCA UNCuyo. Almirante Brown 500. M5528AHB.
Chacras de Coria. Mendoza. Argentina.
3INTA EEA La Consulta. Laboratorio de Fitopatología.
*
dinnocenzo.sandra@inta.gob.ar
Abstract
Crown gall is one
widespread grapevine disease in the world, caused by Allorhizobium vitis (syn.
Agrobacterium vitis) and Agrobacterium tumefaciens (syn. Rhizobium
radiobacter). All. Vitis, is the predominant species and primary
cause of the disease. This study aimed to identify and characterize molecularly
the agrobacteria present in plants with crown gall symptoms in Mendoza
vineyards. Diseased plants with trunk trunk-based galls were sampled from
different areas of Mendoza. For bacterial identification and characterization,
two multiplex PCRs were performed demonstrating that 91.6% of the strains
obtained were agrobacteria (77% A. tumefaciens and 23% All. vitis).
Fifty percent of All. vitis and 16% of A.
tumefaciens identified strains were pathogenic. Pathogenicity tests were
also conducted on Kalanchoe daigremontiana, with resulting tumorigenic
symptoms.
Keywords: Allorhizobium, Agrobacterium,
Vitis vinifera, crown gall, Mendoza
Resumen
Una de las
enfermedades de la vid ampliamente distribuida en el mundo es la agalla de
corona, que tiene como agente causal a Allorhizobium vitis (syn. Agrobacterium
vitis) y Agrobacterium tumefaciens (syn. Rhizobium radiobacter),
siendo la primera especie la que predomina como agente causal de la enfermedad
en vid. El objetivo de este estudio fue identificar mediante técnicas
moleculares las agrobacterias patógenas presentes en plantas con síntomas y
determinar cuál de ellas predomina en viñedos de la provincia de Mendoza.
Plantas de vid con agallas en el tronco provenientes de diversas zonas de la
provincia de Mendoza se utilizaron para realizar los aislamientos. Para la
identificación y caracterización molecular de los aislados se realizaron dos
reacciones múltiples de PCR. Se identificó el 91,6% de las cepas obtenidas como
agrobacterias (77% A. tumefaciens y 23% All. vitis). Se determinó
que el 50% del total identificado como All. vitis son
cepas patógenas, mientras que para A. tumefaciens sólo el 16% de los
aislados dio patogenicidad positiva. También se realizaron pruebas de
patogenicidad en Kalanchoe daigremontiana, donde se observó el
desarrollo de los síntomas típicos de tumorigénesis.
Palabras clave: Allorhizobium, Agrobacterium,
Vitis vinifera, agalla de corona, Mendoza
Originales: Recepción: 03/05/2024 - Aceptación: 04/09/2024
Introduction
With
207,047 hectares cultivated with grapevines (Vitis vinífera), Argentina
leads the international wine industry. The province of Mendoza produces 70% of
Argentinian wine (11) and is considered
one of the Wine Capitals Worldwide. This industry, including grape growing,
wine and must production, and tourism, is fundamental to the economic
development of the province.
Various
pests and diseases significantly reducing production quantity and quality
affect grapevine cultivation. Crown gall, a disease caused by Allorhizobium
vitis (15) and Agrobacterium
tumefaciens (14), is among the most
important and widespread vine diseases globally. These bacteria were first
isolated in the United States in 1907 and were later reported in China, Japan,
South Africa, and some countries in Europe and South America (4).
In
grapevine, All. vitis is the predominant
species causing the disease, while A. tumefaciens is found less
frequently and in smaller proportions. A. tumefaciens is polyphagous and
can affect several dicotyledon species, including Solanaceae and various
Asteraceae (4, 6). Currently, rrs
analysis and constitutive genes have described new species of Agrobacterium initially
identified as A. tumefaciens in various hosts (7).
A.
tumefaciens and All. vitis exist in nature
as pathogenic and non-pathogenic strains. Pathogenic strains contain a
non-essential tumor-inducing plasmid (pTi) involved in disease triggering (16). All. vitis genomic
organization is characterized by two circular chromosomes. The smaller,
chromosome II later classified as a chromid, is essential for disease
development. A. tumefaciens carries one circular chromosome and a
secondary linear chromid (16).
Typically,
the process begins with a wound in the trunk or roots. The wound releases
chemical signals that, perceived by bacteria, induce virulence (2). The disease is triggered when certain genes
from the Ti plasmid are transferred to the host genome, encoding overexpression
of phytohormone synthesis. This overexpression augments cell division
(hyperplasia) and cell size (hypertrophy), leading to the characteristic tumor.
This plasmid also contains genes encoding opine synthesis. Opines are
low-molecular-weight compounds, used by agrobacteria as carbon and nitrogen
source (18). According to Kuzmanović et al. (2020). Ti plasmids are
classified into three major groups: octopine, nopaline and vitopine. Genes
coding for octopine are present in 60% of the strains. About 30% of strains
carry the nos genes (nopaline synthase), and only 10% of strains have
vitopine type (4).
The development of one or more tumors around a diseased organ
alters sap movement, causing chlorosis, vigor loss and decreased production. In
extreme cases, it may lead to plant death, including nursery young plants or
cuttings (4).
Several chromosomal
genes aid in accurate identifications of pathogenic agrobacteria species.
Plasmid genes determine the presence of pathogenicity-related oncogenes. Due to
the importance of viticulture in Mendoza, this study aimed to define the main
molecular traits of the causal agent of crown gall identifying pathogenic
species. We finally aimed to determine the predominant species in Mendoza
vineyards.
Materials
and methods
Plant
samples and Bacterial strains
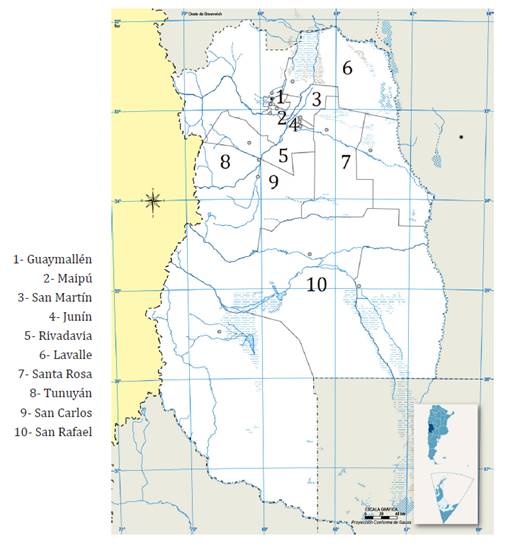
One hundred and forty-eight symptomatic plants (figure
1) were collected from various vine-growing areas of Mendoza (figure 2).
Figure
1. Symptoms of crown gall on Mendoza grapevines.
Figura 1. Síntomas
de agalla de corona en vides de Mendoza.
Figure 2. Mendoza,
sampled departments.
Figura 2. Mendoza,
departamentos muestreados.
Composite
samples were taken from plants within the same vineyard, resulting in 86
samples to be analyzed (table 1).
Table 1. Strain
identification, geographical origin, plant age, cultivar, number of plants,
number of analyzed samples, isolation.
Tabla 1. Identificación
de la cepa, origen geográfico, edad de las plantas, cultivar, número de
plantas, número de muestras analizadas y aislamiento.
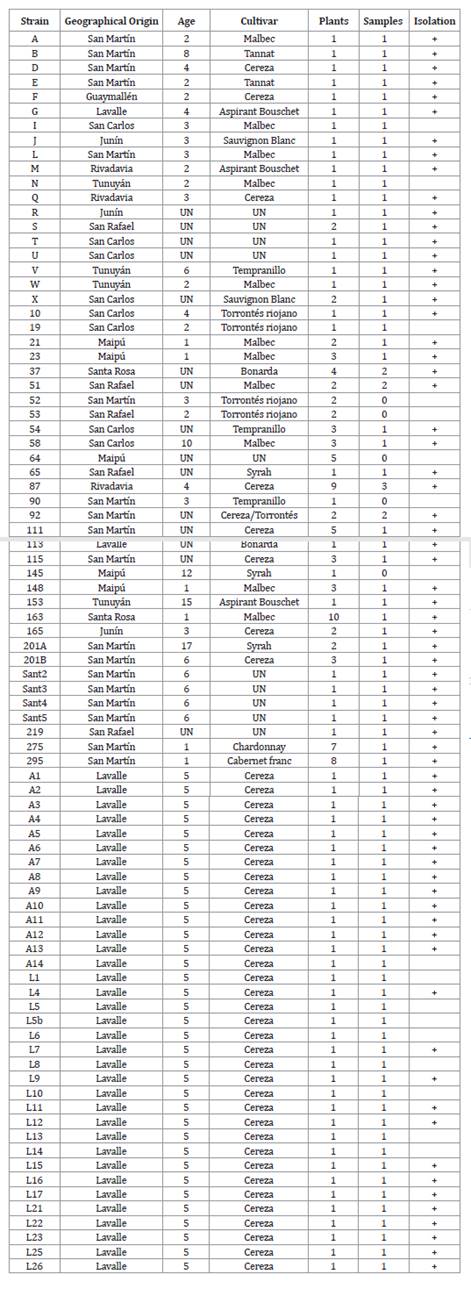
UN:
unknown/desconocido.
Galls were washed
with running water. Subsequently, they were disinfected with 1.1% sodium
hypochlorite for 5 minutes in a laminar flow cabinet. After disinfection, they
were rinsed twice with sterile distilled water, completely removing sodium
hypochlorite. Galls were then cut into small pieces, discarding the external
part to minimize contaminating microorganisms. The resulting pieces were placed
in 5 ml sterile distilled water for one hour allowing diffusion of bacteria in
the sample.
Each bacterial
suspension was streaked onto Roy and Sasser (RS) semiselective culture medium
for All. vitis and Schroth culture medium for A.
tumefaciens. Culture plates were incubated at 27 °C in darkness. Colony
development was observed after seven days. All. vitis
colonies on RS medium had a dark red center with transparent or white
edges. The red center is not always evident (Schaad et
al., 2001 and Burr, T. J. personal
communication, June 28, 2016). A. tumefaciens colonies acquire a reddish
color in Schroth culture medium. Colonies with these characteristics were then
transferred to Luria Bertrani (LB) culture medium.
DNA
extraction and specific PCR amplification
DNA was extracted according to Khlaif, H.
and Al-Karablieh (2002). All. vitis and A.
tumefaciens, were differentiated according to differences in the 23S rDNA
gene (20). A universal forward and two
specific reverse primers were used: B1R for A. tumefaciens and AvR for All.
vitis (table 2).
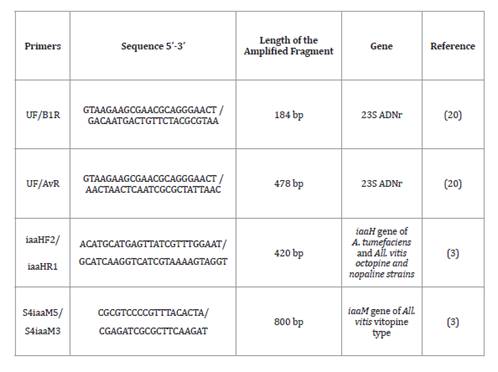
Table 2. Primers
for identification and molecular characterization of All. vitis
and A. tumefaciens strains.
Tabla 2. Primers usados
para identificación y caracterización molecular de cepas de All. vitis y A. tumefaciens.
Multiplex
PCR (polymerase chain reaction) used the following reagents: 1X PCR buffer, 1.5
mM MgCl2, 200 mM dNTP, 1 mM of each primer and 1U of Recombinant DNA polymerase
(Invitrogen) and 5 μl of template DNA for a final reaction volume of 25 μl. The
PCR consisted of initial denaturation at 94°C 1 min, 35 cycles at 94°C 1 min,
67°C 1 min, 72°C 1.5 min and 72°C 10 min, using an Eppendorf thermocycler.
Multiplex
PCR with specific primers for oncogenes allowed for pathogenic strain
detection. The reaction combined the primers iaaHF2/iaaHR1 and S4iaaM5/S4iaaM3
for the auxin-biosynthesis genes iaaH and iaaM, respectively. The reaction was
carried out in a final volume of 25 μl, with 1X Buffer, 1.5 mM MgCl2, 0.5 μM of
each primer, 200 μM of dNTP, 1.25 U of polymerase (Invitrogen Platinum DNA
polymerase) and 1 μl of DNA. Amplification began with initial denaturation at
94°C for 1 min, followed by 30 cycles at 92°C 1 min, 54°C 1 min, 72°C 1.5 min
and 72°C 3 min (3).
PCR-generated
amplicons were detected by electrophoresis using 1% agarose gel, run at 90
volts for 1 hour and stained with ethidium bromide. The gels were visualized
under UV light and photo-documented using Bio-Rad equipment and Quantity One
software. Band size was compared with a 100 bp ladder molecular marker
(Invitrogen).
Pathogenicity tests
PCR results were confirmed via biological tests performed on Kalanchoe
daigremontiana plants to evaluate isolate-pathogenicity. Inoculation was
carried out through punctures on the stem with a micropipette tip and 2.5 μl of
bacterial suspension 109 cfu/ml of each strain. Each strain was inoculated in
three plants via 5 stem wounds per plant. Sterile distilled water was the
negative control and All. vitis and A.
tumefaciens reference strains were positive controls.
The plants were
kept in the laboratory at room temperature, and covered with plastic bags to
maintain approximately 90 % humidity for three days. Then, bags were removed
and plants were taken to the greenhouse. Observations were made every 15 days
for two months (23). Isolations from
tissues developed in the inoculation zone were carried out in a semiselective
culture medium, using the same method as with vine galls.
Results
Molecular
analysis
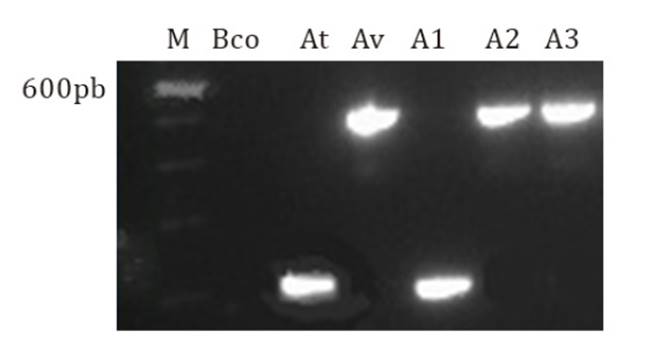
Sixty-nine isolates out of 86 samples analyzed resulted in 91.6%
identified as agrobacteria, among which 77% were A. tumefaciens and 23%.
All. vitis. Figure 3 presents
a PCR with 3 isolates where A1 was A. tumefaciens and A2 and A3 were All.
vitis.
From left to right: M: marker 100 bp (PROMEGA); Bco:
water; At: A. tumefaciens reference strain; Av: All. vitis reference strain.; gall isolates: A1, A2, A3.
De izquierda a
derecha: M: marcador 100 pb (PROMEGA); Blanco: agua; At: cepa de referencia de A.
tumefaciens; Av: cepa de referencia de All. vitis;
aislados de agalla: A1, A2, A3.
Figure
3. Multiplex PCR with primer pairs UF/B1R (184 bp) and
UF/AvR (478 bp).
Figura 3. PCR
múltiple con los pares de primers UF/B1R (184 pb) y UF/AvR (478 pb).
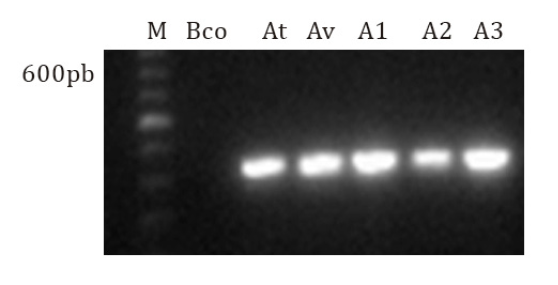
The multiplex PCR was performed with the combination of primers
iaaHF2/iaaHR1, while S4iaaM5/S4iaaM3 determined pathogenicity. The iaaH gene
was only amplified on 16% of A. tumefaciens strains, molecularly
identified as pathogenic, and 50% of All. vitis isolates
proved to be pathogenic. The iaaM gene did not amplify. Figure
4 shows amplification of the pathogenicity gene present in both species.
Isolates A1 of A. tumefaciens and A2 and A3 of All. vitis present positive pathogenicity.
From left to right: M: 100bp marker (Promega), Bco:
water; At: A. tumefaciens reference strain; Av: All. vitis reference strain; gall isolates: A1, A2 and A3.
De
izquierda a derecha: M: marcador 100 pb (Promega), Blanco: agua; At: cepa de
referencia de A. tumefaciens; Av: cepa de referencia de All. vitis; aislados de agalla: A1, A2 y A3.
Figure
4. Multiplex PCR with primer pairs iaaHF/iaaHR (420 bp)
and S4iaaM5/S4iaaM3 (800 bp).
Figura 4. PCR
múltiple con los pares de primers iaaHF/iaaHR (420 pb) y S4iaaM5/S4iaaM3
(800 pb).
Pathogenicity
Test
Two weeks after inoculation, positive results were observed in Kalanchoe
plants. Abnormal growth and color change (redness) were similar to those in
plants inoculated with reference strains. In some cases, corky tissue developed
at the inoculation site (figure 5). These results became
more pronounced two months after inoculation.
A: Negative control (water), B: reference strain of A.
tumefaciens, C: reference strain of All. vitis,
D, E and F: gall isolates: A1, A2 and A3 with positive pathogenicity result
(molecular analysis).
A: Control
negativo (agua), B: cepa de referencia de A. tumefaciens, C: cepa de
referencia de All. vitis, D, E y F: aislado
muestra con resultado de patogenicidad (análisis molecular) positivo.
Figure
5. Symptoms observed in Kalanchoe stems 2 weeks
after inoculation.
Figura 5. Síntomas
observados en tallos de Kalanchoe a las 2 semanas de la inoculación.
Discussion
Bacterial genetic diversity of both species limits detection
efficiency in grapevines (3). All. vitis strains are genetically diverse (4; 8; 9; 16; 23).
Our data suggest All. vitis could be a species
complex comprising several genomic species (16).
In this study, after obtaining pure and simple isolates, successful species
identification followed the molecular protocol described by Pulawska et al. (2006). However, since this PCR
does not identify pathogenicity genes, the analysis must be complemented with
additional PCR determining gene presence (1, 3, 8,
17, 19, 22). This research used specific primers iaaHF2/iaaHR1 and
S4iaaM5/S4iaaM3 for iaaH and iaaM genes, respectively, showing non-pathogenic A.
tumefaciens strains predominated over All. vitis
strains in the analyzed grapevine samples. However, 50% of All. vitis isolates were pathogenic. This finding indicates
that All. vitis is the predominant pathogenic
species and main disease cause in grapevines studied in Mendoza, in agreement
with prior studies (2, 5, 10, 14, 22).
The same PCR
determining pathogenicity, determined opine type. We found absent vitopina type
in all All. vitis isolates and
presence of the octopine/nopaline types in Mendoza,
Seventy-nine
percent of pathogenicity tests in inoculated Kalanchoe showed disease
symptoms. This value is within the expected range (78-94%) (21). These data also align with Kuzmanović et al. (2016), who observed that some
strains did not demonstrate their tumorigenic capacity in inoculated plants
despite possessing pathogenicity-associated genes molecularly identified. This
suggests that such isolates remain potentially tumorigenic. However, pathogenicity
is influenced by plant age and environmental conditions. Absent Crown gall
symptoms do not imply absent tumorigenesis genes (18),
probably because no single host is infected by more than 81% of pathogenic
strains and not all strains produce tumors in every host (13). According to Lamovšek
et al. (2014), determining pathogenicity through molecular tests
might replace biological tests. The PCR are less time-consuming and
labor-intensive. However, given the occurrence of false negatives,
pathogenicity tests remain a valuable tool in plant bacteriology.
Conclusions
This study
successfully identified and characterized the causal agents of Crown gall in
Mendoza vineyards using molecular methods. Our methodology enables the
characterization of agrobacteria in Argentina and provides a quick and precise
diagnostic tool, even for evaluating asexually propagated grapevines.
This information
will help develop management strategies to reduce disease spread and incidence
in our vineyards and nurseries and improve the health and productivity of their
vineyards.
Finally, our results
aiding bacterial identification in plant material allow for protocols to detect
bacteria in asymptomatic material ensuring propagation of healthy plants from
health-controlled material.
To the best of our knowledge, this is the first study identifying
uncited crown gall species in Argentina.
Acknowledgment
Dr. Ibrahim Tolba (Plant Pathologist at the Faculty of
Agriculture, Ain Shams University, Egypt) for supplying positive controls.
1. Alippi, A. M.;
López, A.; Balatti, P. 2011. Métodos para la detección de Agrobacterium a
partir de muestras de material vegetal, suelo y agua. Revista Argentina de
Microbiología. 43: 278-286.
2. Argun, N.;
Maden, S. Momol, M. T.; Momol, E. A.; Reid, C. L.; Celik, H.; Burr, T. J. 2002.
Characterization of Agrobacterium vitis strains isolated from Turkish
grape cultivars in the Central Anatolia Region. Plant Disease. 86(2): 162-166.
3. Bini, F.;
Kuczmog, A.; Putnoky, P.; Otten, L.; Bazzi, C.; Burr, T. J.; Szegedi, E. 2008.
Novel pathogenspecific primers for the detection of Agrobacterium vitis and
Agrobacterium tumefaciens. Vitis. 47(3): 181-189.
4. Burr, T. J.;
Bazzi, C.; Süle, S.; Otten, L. 1998. Biology of Agrobacterium vitis and
the development of disease control strategies. Plant Disease. 82(12):
1288-1297.
5. Canik Orel, D.;
Karagoz, A.; Durmaz, R.; Ertunc, F. 2016. Phenotypic and molecular
characterization of Rhizobium vitis strains from vineyards in Turkey.
Phytopathologia Mediterranea. 55(1): 41-53. DOI:
10.14601/Phytopathol_Mediterr-15776
6. Chen, F; Hseu,
S.; Hung, S.; Chen, M.; Lin, C. 1999. Leaf, stem and crown galls on perennial
asters caused by Agrobacterium tumefaciens in Taiwan. Bt. Bull. Acad.
Sin. 40: 237-242.
7. Flores-Félix,
J.; Menéndez, E.; Peix, A.; García-Fraile, P.; Velázquez, E. 2020. History and
current taxonomic status of genus Agrobacterium. Systematic and Applied
Microbiology. 43(1):126046. https://doi.org/10.1016/j.syapm.2019.126046
8. Genov, I.;
Atanassov, I.; Tsvetkov, I.; Atanassov, A. 2006a. Isolation and
characterization of Agrobacterium strains from grapevines in Bulgarian
vineyards and wild grapes, V. vinifera ssp. Silvestris. Vitis. 45(2):
97-101.
9. Genov, I.;
Atanassov, I.; Yordanov, Y.; Tsvetkov, I.; Atanassov, A. 2006b. Genetic
diversity of Agrobacterium vitis strains, isolated from grapevines and
wild grapes in Bulgaria, assessed by Cleaved Amplified Polymorphic Sequences
analysis of 16S-23S rDNA. Vitis. 45: 125-130.
10. Genov, N.; Llop, P.; López, M.; Bobev, S.; Álvarez, B. 2015.
Molecular and phenotypic characterization of Agrobacterium species from
vineyards allows identification of typical Agrobacterium vitis and
atypical biovar 1 strains. Journal of Applied Microbiology. 118(6): 1465-1477.
DOI: 10.1111/jam.12791
11. INV. 2022.
Informes anuales.
https://www.argentina.gob.ar/inv/vinos/estadisticas/superficie/ anuarios
12. Khlaif, H.;
Al-Karablieh, N. 2002. Occurrence and distribution of crown gall disease in
Jordan. Phytopathology Mediterranean. 41: 226-234.
13. Kuzmanović, N.;
Gašić, K.; Ivanović, M.; Prokić, A.; Obradović, A. 2012. Identification of Agrobacterium
vitis as a causal agent of grapevine crown gall in Serbia. Archives of
Biological Sciences. 64(4): 1487-1494. DOI:10.2298/ABS1204487K
14. Kuzmanović, N.;
Biondi, E.; Ivanović, M.; Prokić, A.; Zlatković, N.; Bertaccini, A.; Obradović,
A. 2016. Evaluation of different PCR primers for identification of tumorigenic
bacteria associated with grapevine crown gall. Journal of Plant Pathology. 98(2):
311-319.
15. Kuzmanović, N.;
Biondi, E.; Overmann, J.; Puławska, P.; Verbarg, S.; Smalla, K.; Lassalle, F.
2020. Revisiting the taxonomy of Allorhizobium vitis (i.e.
Agrobacterium vitis) using genomics -emended description of All. vitis sensu stricto and description of Allorhizobium
ampelinum sp. nov. bioRxiv.
https://doi.org/10.1101/2020.12.19.423612
16. Kuzmanović, N.;
Biondi, E.; Overmann, J.; Puławska, P.; Verbarg, S.; Smalla, K.; Lassalle, F.
2022. Genomic analysis provides novel insights diversification and taxonomy of Allorhizobium
vitis (i.e. Agrobacterium vitis). BMC Genomic 23: 462. https://doi.
org/10.1186/s12864-022-08662-x
17. Lamovšek, J.;
Zidarič, I.; Mavrič Pleško, I.; Urek, G.; Trdan, S. 2014. Comparative study of
diagnostic methods used for monitoring of common grape vine (Vitis vinifera L.)
crown gall (Agrobacterium vitis Ophel & Kerr) in Slovenia.
Acta Agriculturae Slovenica. 103.
18. Llop, P. P.
2003. Caracterización molecular de la pérdida del poder patógeno en Agrobacterium
tumefaciens. Tesis doctoral. Facultad de Ciencias Biológicas. Universidad
de Valencia. España.
19. Peduto, F.;
Marchi, G.; Surico, G. 2010. Indexing Agrobacterium vitis in asymptomatic
grapevine propagation material by two nested PCR Assays. American Journal of
Enology and Viticulture. 61(1): 102-112.
20. Pulawska, J.;
Willems, A.; Sobiczewski, P. 2006. Rapid and specific identification of four Agrobacterium
species and biovars using multiplex PCR. Systematic and Applied
Microbiology. 29: 470-479.
21. Schaad, N. W.;
Jones, J. B.; Chun, W. 2001. Laboratory guide for identification of plant
pathogenic bacteria, 3° Edición. Minnesota.
22. Szegedi, E.;
Bottka, S. 2002. Detection of Agrobacterium vitis by polymerase chain
reaction in grapevine bleeding sap after isolation on a semiselective medium.
Vitis. 41: 37-42.
23. Tolba, I.; Zaki, M. 2011. Characterization of Agrobacterium
vitis isolates obtained from galled grapevine plants in Egypt. Annals of
Agricultural Science. 56: 113-119. doi:10.1016/j.
aoas.2011.06.001