Revista de la Facultad de Ciencias
Agrarias. Universidad Nacional de Cuyo. En prensa. ISSN (en línea) 1853-8665.
Original article
Epiphytic
microorganisms associated with banana phyllosphere with potential antagonism to
Black Sigatoka (Pseudocercospora fijiensis) in Los Ríos, Ecuador
Identificación
de microorganismos epífitos asociados a la filósfera de banano con potencial
antagonismo a Sigatoka Negra (Pseudocercospora fijiensis) en la
provincia de Los Ríos, Ecuador
Solanyi Marley Tigselema Zambrano1*,
Aracelly Mabel Villalba Puga2,
Jim Raphael Ochoa Ramos1,
Galo Efraín Lara Hidalgo1,
Diana Aracelly López1
1 Instituto Nacional de Investigaciones Agropecuarias. Mocache.
Ecuador. C. P. 120310.
2 Universidad de Las Américas. Facultad de Ingeniería y Ciencias
Aplicadas. Quito. Ecuador. C. P. 170124.
* solanyi.tigselema@iniap.gob.ec
Abstract
Black Sigatoka (Pseudocercospora
fijiensis) is the most important leaf spot disease of bananas worldwide,
particularly affecting Cavendish banana, the most exported variety.
Additionally, this pathogen has developed resistance to some effective fungicides,
making its management increasingly difficult. Epiphytic microorganisms with
potential antagonism to P. fijiensis were identified in conventional
banana farms in the province of Los Ríos. Sampling areas were determined
through zoning processes and selecting the cantons of Mocache, Valencia, Baba
and Pueblo Viejo. Leaf tissue samples were collected from three farms per zone.
Microorganisms were isolated and morphologically and molecularly characterised
in nine farms in the cantons of Valencia (63 bacteria), Baba (39 bacteria),
Pueblo Viejo (8 bacteria) and 8 genera of fungi including 15 species. The
isolated bacteria presented macroscopic and microscopic characteristics with
different shapes, elevations, edges, consistencies and pigmentations. Taxonomically,
they belonged to the genera Bacillus and Cocos, 81% Gram-negative
and 19% Gram-positive. The analysis conducted for sampling-site selection
allowed the identification of different microbial behaviours.
Keywords: microorganisms,
fungi, bacteria, Musa spp.
Resumen
Sigatoka negra es
la enfermedad de la mancha foliar más importante del banano a nivel mundial. La
variedad de banano Cavendish, es considerada la más común y la más exportada;
sin embargo, presenta una alta susceptibilidad frente a la enfermedad. Existen
fungicidas altamente efectivos para su control; sin embargo, el patógeno ha
logrado generar resistencia a algunos de estos, lo que ha dificultado cada vez
más su manejo. Se identificaron microorganismos epífitos con potencial
antagonismo a Pseudocercospora fijiensis en fincas de banano
convencionales de la provincia de Los Ríos. Las zonas de muestreo fueron
determinadas a través de procesos de zonificación, seleccionando los cantones
Mocache, Valencia, Baba y Pueblo Viejo. Se recolectó muestras de tejido foliar
en tres fincas por zona. Se aislaron microorganismos y se caracterizaron
morfológica y molecularmente en nueve fincas en los cantones de Valencia (63
bacterias), Baba (39 bacterias) y Pueblo Viejo (8 bacterias) y 8 géneros de hongos
que incluyen 15 especies. Las bacterias aisladas presentaron características
macroscópicas y microscópicas con diversas formas, elevaciones, bordes,
consistencias y pigmentaciones, así como diversas taxonomías pertenecientes a
los géneros Bacillus y Cocos, siendo 81% Gram negativas y 19% Gram
positivas. El análisis realizado para la selección de los sitios de muestreo
fue apropiado ya que se observó un comportamiento diferencial de los
microorganismos en estas zonas.
Palabras claves: microorganismos, hongos,
bacterias, Musa spp.
Originales: Recepción: 21/08/2024
- Aceptación: 06/03/2025
Introduction
Musaceae is a family of
monocotyledonous plants that include bananas and plantains, often called giant
herbs (31). These plants belong to the genus Musa,
cultivated in tropical and subtropical regions (36).
The banana sector
in Ecuador has 167,893 hectares, with a productivity of 6,684,916
tons. Los Ríos province has the highest participation in the national
production of fresh fruit with 38.47% (2 571 356 t), with a contribution of 1
328 537 964.98 US dollars (45).
Investment in
production and related industries (goods and services needed for banana
production) and the current banana export process created jobs for more than
one million households in Ecuador, benefiting around 2.5 million people in nine
provinces heavily dependent on the banana industry. Compared to other non-oil
sectors in the country, this sector is the backbone of economic activity,
generating higher incomes and providing more employment opportunities (16).
Black Sigatoka is
the most economically important leaf spot disease of Musaceae, affecting
many plantations and resulting in forced early harvesting (27). This disease is
caused by the fungus Pseudocercospora fijiensis, exclusive of banana
foliage with sexual and asexual reproduction. It infects the plants, hindering
photosynthesis and causing gradual leaf necrosis and death. Disease severity is
determined by the Stover scale modified by Benavides-López (2019), Gauhl
(1994)
and Muimba-Kankolongo
(2018).
Black Sigatoka is
mainly controlled by technical management and appropriate fungicide rotation.
However, given climatic variability, the disease shows different behaviours
around the country. Los Ríos province is the most affected, with 74% of
production losses. Twenty-two to 29 annual aerial spraying cycles are used to
fight the disease, representing costs between $430 and $800 (8,
9). In addition, surgical practices like excision of mottled areas
and leaf removal are carried out (9, 17).
International
markets for plant protection products are dominated by synthetic pesticides (30). These chemical
substances seriously affect the ecosystem and induce resistance, altering
ecological equilibriums (28). Therefore,
searching for alternative control strategies is relevant worldwide (24).
The search for
antagonistic microorganisms for biological control of pathogens in economically
important crops has aroused particular interest due to their potentialities (4,
54). Microorganisms of agricultural importance represent a key ecological
strategy towards the integrated development of practices such as nutrient,
disease and pest management, reducing chemicals and improving crop yield (42).
Several microorganisms showing beneficial effects on plants may constitute
potential biocontrol agents (3, 41) and important
actors in sustainable agriculture (51).
Biological control
of Black Sigatoka has received relatively little attention due to the
availability of highly effective fungicides. However, the emergence of pathogen
isolates resistant to systemic fungicides and the need for cleaner production
technologies have increased interest in biological control (22).
The search for
effective biological products against this disease has studied different
microorganisms associated with these crops (12). Therefore, this
research aimed to collect, isolate and characterise microorganisms from the
phyllosphere of Musaceae.
Materials
and methods
Study
area
This research was
conducted at the Pichilingue Tropical Experimental Station (EETP) of the
National Institute of Agricultural Research (INIAP) with samples obtained from
conventional banana farms of Cavendish Williams cultivar in the cantons of
Mocache, Valencia, Baba and Pueblo Viejo, Los Ríos province.
Field
methodology
Sampling sites were
chosen by zoning with cartographic charts from the IGM (Military Geographic
Institute) database. The climate micro-zonation map was generated by satellite
images with ArcGIS 10.8, at a scale of 1:25 000 for geo pedological conditions
considering soil pH, organic matter and surface texture, with 1:50 000 scale,
considering geopedology, geomorphology, CUT (soil usage capacity) and isotherms,
including climatic zones, temperature and cover use (figure 1).
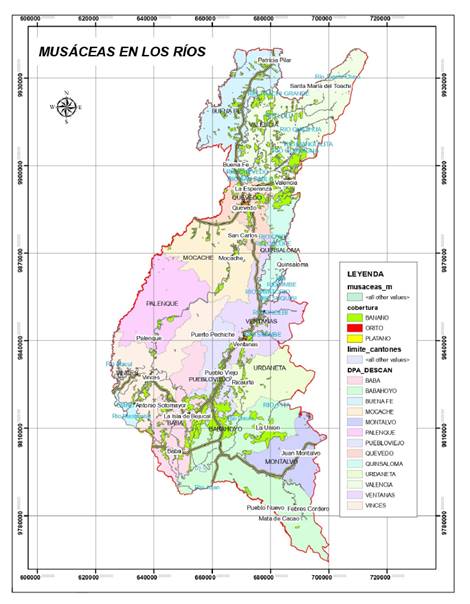
Figure 1. Climate
micro-zonation map for sampling sites in the Province of Los Ríos, generated by
ArcGIS 10.8 software.
Figura
1. Mapa de microzonificación climática
para sitios de muestreo en la Provincia de Los Ríos, generado con el software
ArcGIS 10.8.
Ten subsamples were
collected from each farm, constituting one composite sample. In selected
plants, the third and fourth leaves were identified for tissue to be obtained
from the central third, both on the right and left side of the midrib. Samples
were identified by recording origin and date (41).
Fungal
identification from leaf tissue was conducted in Mocache, Baba and Pueblo
Viejo. Bacteria were identified from leaves in Valencia, Baba and Pueblo Viejo.
Microorganism
isolation
For the isolation
of bacteria, the samples obtained were processed according to Intriago
Mendoza (2010). Agar culture medium was prepared in flasks, sterilised in autoclave
for 30 minutes and distributed in petri dishes. Twenty-five g of leaf tissue
were washed in 100 mL of sterile distilled water (SDW). Product water was used
for serial dilutions up to -3. One ml of each dilution was seeded by Digralsky
loop, and plates were incubated at room temperature for 5 days for growth
evaluation.
In order to isolate
fungi, plant tissue samples were washed with distilled water, cut into small
portions of tissue (3 to 5 mm) and immersed in a 1-3% hypochlorite solution for
one minute, followed by rinsing with sterile water. Tissue portions were seeded
in Petri dishes with PDA (potato dextrose agar) + chloramphenicol medium and
incubated at 28°C for 5 days. Isolates were purified and preserved at 5°C (20).
Morphological
characterization
After biochemical
Gram staining and catalase tests, macroscopic and microscopic characterisation
was carried out on the isolated microorganisms, described by their shape,
colour, edges, elevation and consistency (52).
Number
of Colonies
Number of colonies on the plates is expressed as CFU/ml (Colony
Forming Units) according to Casas et al. (2017).
Extraction
of fungal genomic DNA
According to Doyle & Doyle (1987) modified by Faleiro
et al. (2002), samples were split into two boxes per sample with 14 daysqold
mycelium and triturated with liquid nitrogen. The homogenate was mixed with 800
μL extraction buffer (7% cetyltrimethylammonium bromide [CTAB], 5 M NaCl, 0.5
mM ethylenediaminetetraacetic acid [EDTA], 1 M Tris-HCl pH = 8,
Polyvinylpyrrolidone (PVP-40), 1% (v/v) ß-mercaptoethanol and milliQ water).
Five μL of proteinase K (concentration 20 mg/uL) was added to the homogenate
and incubated at 65°C for 1 hour in a water bath. Then, it was centrifuged at
14 000 rpm for 15 minutes and the supernatant was collected into 2 mL tubes,
added 55 μL of 7% CTAB and 700 μL chloroform: isoamyl alcohol (25: 1; v: v),
mixed with inversion and vortexed until an emulsion was formed and centrifuged
at 14 000 rpm for 16 minutes. Again, the supernatant was extracted to 2 mL
tubes by adding 700 μL chloroform: isoamyl alcohol (25: 1; v: v), mixed and
centrifuged. The supernatant was recovered in 1.5 mL tubes and 700 μL (2/3 of
the tube) was added with ice-cold absolute ethanol (-20°C), for storage at
-20°C for 1 to 2 hours. Centrifugation was performed at 14 000 rpm for 5
minutes, obtaining a white pellet and the supernatant was removed by washing
the pellet 3 to 4 times with 70% ethanol at -2°C. (500 μL). Finally, the pellet
was dried in a thermoblock at 55°C and DNA was resuspended in 100 μL of TE with
RNAsa (concentration 20 mg/ml).
PCR
Amplification of Ribosomal ITS Region
To verify the
extracted DNA, amplification was performed with markers ITS1
(TCCGTAGGTGAACCTGCGG) and ITS4 (TCCTCCGCTTATTGATATATGC) and the amplification
cocktail proposed by Morillo & Miño (2011). All samples were
amplified on the Applied Biosystems thermal cycler in a total reaction volume
of 25 μl, including 2.50 μL of 5x Green GoTaq® Flexi Buffer (Promega), 1.5μL of
MgCl2 (25 mM), 0.50 μL of dNTPs (5mM), 2 μL of each primer (5 μmol), 0.50 μL of
DNA polymerase (Thermo Scientific DreamTaq) (5 U/ μl), 1 μL of genomic DNA (5
ng/ μl) and 15 μL of ultrapure water. PCR included initial denaturation at 95°C
for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min,
hybridisation at 55°C for 2 min, elongation at 72°C for 1 min and a final
elongation step at 70°C for 10 min.
DNA amplicons were
analysed on a 1.5% (w/v) agarose gel using Syber Safe for 30 minutes at 100V.
Amplicon sizes were estimated by comparison with a TrackIt™ 1 Kb Plus DNA
Ladder molecular weight marker and visualised in a photodocumenter.
Sequence
analysis
PCR products were
shipped to the research laboratories of the Universidad de las Américas (UDLA),
according to the university guidelines, which consisted of 10 μL of PCR
product, 2 μL of each primer (ITS 1/ ITS4) (2 μM concentration) per sample and
cold chain storage at 4°C or below. Sequence editing was performed using the
Unipro UGENE software and the BLAST programme at the Centro Nacional de
Información Biotecnológica (http://www.ncbi.nlm.nih.gov/) to obtain consensus
sequences. Sequence alignment was performed by MUSCLE algorithm with MegaX
software. The matrix obtained was used to assemble the phylogenetic tree based
on the Maximum-Likelihood (ML) algorithm, with genetic distance calculated by the
Jukes-Cantor model, Bootstrap-100 and Uniform Rates.
Results
Colony
count
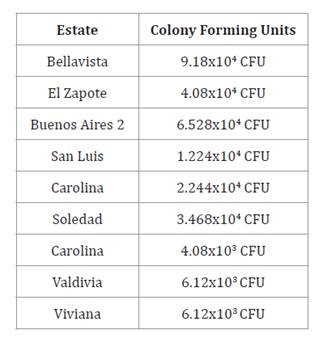
Considering the three farms evaluated in each canton, in
Valencia, the Bellavista farm had the highest number of colonies with 9.18x104
CFU; in Baba the Soledad farm had the highest number of colonies
with 3.468x104 CFU and in Pueblo Viejo,
Valdivia and Viviana farms had 6.12x103 CFU,
obtaining a total of 63 bacteria in Valencia, 39 bacteria in Baba and 8
bacteria in Pueblo Viejo.
Strain
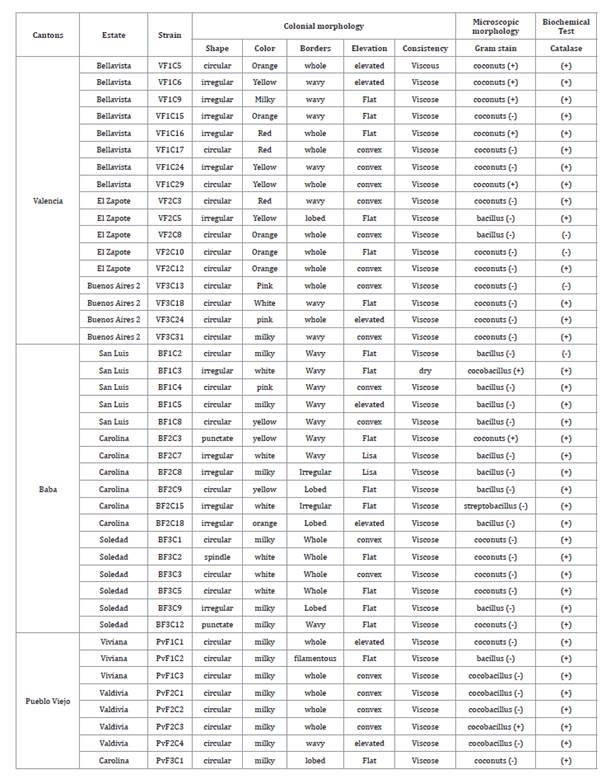
characterisation
Colonial morphological characterisation, microscopic morphology
and biochemical tests were performed on the bacteria isolated in the province
of Los Ríos (table
1).
Table 1. Characterisation
of microorganisms associated with the banana phyllosphere, Los Ríos province.
Tabla
1. Caracterización de microorganismos
asociados a la filósfera de banano, provincia de Los Ríos.
Eight fungal strains were isolated in Mocache, 5 in Baba and 13
in Pueblo Viejo. Eight PCR products were amplified from Mocache, 5 from Baba
and 13 from Pueblo Viejo. Twenty-six sequences were obtained. Similarity
percentages were 90-100 % with NCBI homologous sequences (table 2).
Table 2. Collection
and ITS rDNA sequences of fungi isolated in the cantons of Mocache, Baba and
Pueblo Viejo.
Tabla
2. Detalles de la colección y
secuencias ITS del ADNr de hongos aislados en los cantones de Mocache, Baba y
Pueblo Viejo.
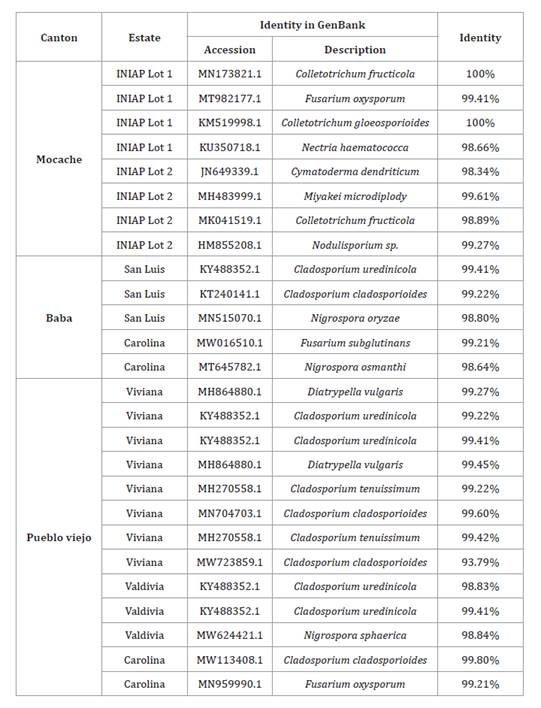
The identified fungi were from the genera Fusarium sp., Colletotrichum
sp., Diatrypella sp., Nodulisporium sp., Nigrospora sp.,
Microdiplodia sp., Cymatoderma sp. and Cladosporium
sp. Fusarium sp. is a candidate biological
control agent against Pseudocercospora fijiensis. Nodulisporium sp. is an endophyte capable of producing insecticidal
nodilosporic acids and volatile antifungal substances (Suwannarach
et al., 2013). Nigrospora sp. produce
bioactive secondary metabolites with antifungal activity (23) (figure 2).
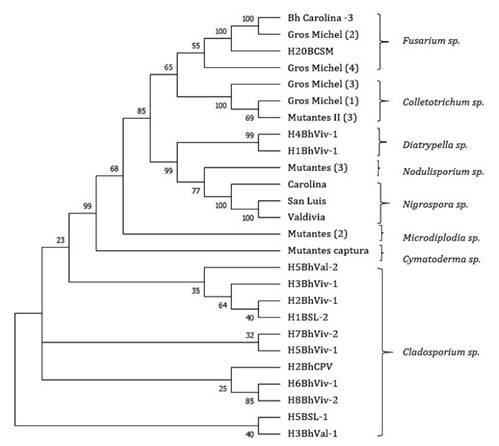
Figure 2. Phylogenetic
tree of fungal isolates associated with banana phyllosphere with potential
antagonism to Black Sigatoka in the cantons of Mocache, Baba and Pueblo Viejo.
Phylogenetic construction by Maximum- Likelihood method, Jukes-Cantor
model, Bootstrap-100 and Uniform Rates.
Figura
2. Árbol filogenético de aislamientos
de hongos asociados a la filósfera de banano con potencial antagonismo a
Sigatoka Negra en los cantones de Mocache, Baba y Pueblo Viejo. Construcción filogenética
por el método Maximum-Likelihood, Jukes-Cantor model, Bootstrap-100 y Uniform
Rates.
Discussion
Fungal disease
control mainly relies on the application of agrochemicals. However, this
practice causes pathogen resistance after prolonged application, generating
public concern about the effects of toxic residues on human health and the
environment.
This research identified Bacillus bacteria. According to Cruz-Martín
et al. (2018) Bacillus pumilus CCIBP-C5 decreases fungal biomass,
induces phytodefense mechanisms in the plant, and may constitute a potential
biological control agent against P. fijiensis. Based on this finding, B.
pumilus CCIBP-C5 constitutes a guideline for further research.
Contrary to other
studies, proportions of Gram-negative bacteria (81%) exceeded that of
Gram-positive bacteria (19%). Previously, Ceballos et al. (2012) found higher
proportions of Gram-positive bacteria (67%) than Gram-negative bacteria from
three banana and plantain cultivars in Urabá (Northwest Colombia). Regarding
the inhibitory capacity of the isolated microorganisms, the results showed that
Bacillus bacteria have antagonistic activity against the fungus P.
fijiensis, as seen by Villegas-Escobar et al. (2013) when isolating 649
strains of aerobic endospore-forming bacteria. The strain Bacillus subtilis showed
the highest inhibition (89±1%), proving its bioactive potential against P.
fijiensis.
After isolation,
colonies showed different shapes (circular, pointed, irregular, spindle),
elevations (flat, convex, smooth, raised), edges (entire, wavy, lobulated,
irregular, filamentous), consistencies (viscous, dry) and pigmentations
(orange, yellow, red, pink, milky, white), as reported by Alfaro
(2013)
when isolating and quantifying epiphytic bacteria from the banana phylloplane Musa
AAA cv. Grande Naine.
Considering the 26
isolated fungi, 25 belonged to Ascomycota and one to Basidiomycota. Ascomycota
fungi grow in subtropical conditions, as bananas (44).
Fungal molecular
identification is frequently assessed with the internal transcribed spacer
(ITS) region (47). The primers ITS1
and ITS4 have broad utility and presence in universal databases, with
successful amplification rates of fungal lineages (53). Based on in
silico analysis (49) the primer ITS1 represents
73.8% of Ascomycota and 85.6% of Basidiomycota in the SSU region, while the
primer ITS4 represents 97.6% in Ascomycota and 96.9% in Basidiomycota in the
LSU region. This means ITS primers allowed amplifying sequences from both
Ascomycota and Basidiomycota fungi.
In a study of
pathogenic taxa in wild banana (Musa acuminata), Brown et
al. (1998) identified potential endophytic pathogen genera and species
that may remain dormant, such as Colletotrichum sp. and
Nigrospora sp. Zakaria & Aziz (2018) isolated fungi of
the genus Nigrospora sp., Fusarium sp. and
Colletotrichum sp. on bananas. Very similar
results were obtained by Horra (2014) and in the present study, including Diatrypella
sp., Nodulisporium sp., Microdiplodia sp., Cymatoderma sp.
and Cladosporium sp.
Fusarium oxysporum is present among
the rhizosphere microflora, and some strains cause wilting or total root rot of
banana plants (19). It should be
noted that all F. oxysporum strains are saprophytes, surviving for long
periods in both soil organic matter and the rhizosphere. This possibly explains
their latent presence in leaf tissue and the sampled areas. We also isolated Nectria
haematococca (sexual morph of F. solani), a filamentous type of
fungus. F. solani is part of a complex with 60 phylogenetic species (46). These two Fusarium
sp. strains could constitute candidate biological
control agents against P. fijiensis (2), encouraging
future antagonistic tests for evaluations against black Sigatoka.
Colletotrichum sp. predominates in the tropics and subtropics with heavy
rainfall and high relative humidity (43). Two species were
found in this study, C. gloeosporioides and C. frutícola. The
former causes banana anthracnose and leaf spot (38), while C. frutícola
has been identified on mango plants (29). One of the main
characteristics of Colletotrichum sp. infection
in bananas is the difficulty in detecting the disease before fruit generation,
given latency (37).
Diatrypaceae members like the
genus Diatrypella are saprobes and pathogens associated with different
hosts in both terrestrial and aquatic environments (14). Species of this
genus are separated according to their entostrophic morphology and the number
of ascospores they present (50). Fungal
pathogenicity is moderate for this genus. This study firstly reports Diatrypella
vulgaris, in banana. However, it is commonly isolated from diseased Vitis
vinifera, causing necrotic lesions on the plant (40).
The fungal isolate Nodulisporium
sp. produces nodilosporic acids with insecticidal
properties and volatile antifungal substances used against other microorganisms
through mycofumigation (48).
Fungi of Nigrospora sp. are
host-specific phytopathogenic, endophytic and saprophytic species that produce
bioactive secondary metabolites with antifungal activity (23). During
isolations, N. sphaerica and N. osmanthi were isolated from the
environment. N. sphaerica exhibits a violent spore-discharging mechanism
that projects spores over long distances (57). Similarly, Nigrospora
oryzae and N. sphaerica were identified from banana leaves (58). This study also
firstly reports N. osmanthi in bananas.
Isolated fungi of
the genus Microdiplodia sp. and Cymatoderma sp.
have not previously been recorded in bananas. However,
Pinheiro
da Costa et al. (2021) detail the presence of Microdiplodia sp. with antifungal activity on Brugmansia suaveolens and Cymatoderma
sp. as the only Basidiomycota reported in tropical
rainforest (1).
Finally, the genus Cladosporium
sp. comprises more than forty species, including
pathogenic species causing leaf spot and saprophytic species acting on
vegetation and soil (35). Isolated C.
cladosporioides confirms this fungus as an endophyte isolated from foliar
culture tissues of banana plants (58). In addition, C.
uredinicola, also isolated in this study, presents small conidia formed
with branched chains that facilitate its propagation over long distances (6), explaining its
higher prevalence over other fungi. Another species identified, C. tenuissimum
is an abundant saprobe in the tropics (56). Moubasher
et al. (2016) state that the frequency of Cladosporium sp. isolation is moderate and peaks during winter, when humidity
benefits banana development.
Conclusions
Considering the
differential behavior of microorganisms and the number of strains found in the
studied sites, the analysis focused on selecting sampling sites was
appropriate.
The isolated microorganisms presented macroscopic and
microscopic characteristics with different shapes, elevations, borders,
consistencies and pigmentations. Different taxonomies belonged to the Bacillus
and Coccus genera, 81% being Gram-negative and 19% Gram-positive.
Acknowledgements
The authors thank all the technical staff of the National
Agricultural Research Institute, who provided their scientific contribution
during the development of the research.
1. Agudelo, M. B.;
Calderón, M.; Betancourt, O.; Gallejo, Á. S. 2007. Hongos Macromycetes en dos
rellictos de bosque húmedo tropical montano bajo de la vereda La Cuchilla.
Marmato, Caldas. Museo de História Natural. 11: 19-31.
2. Alfaro, F. 2013.
Aislamiento y cuantificación de bacterias epífitas del filoplano de banano
(Musa AAA cv. Grande Naine) y selección de cepas quitinolíticas y
glucanolíticas como potenciales antagonistas de Mycosphaerella fijiensis,
agente causal de la Sigatoka negra.
3. Ayala, D.;
Acevedo, C.; Enrique, B.; Aguilar, F.; Orlandoni, G. 2018. Seguimiento de la
actividad PGPR de microorganismos inoculados en plantas de Sacha inchi (Plukenetia
volubilis) bajo condiciones de vivero. Archivo digital de prácticas,
Microbiología Industrial. Universidad de Santander UDES. 1-14.
https://doi.org/10.13140/RG.2.2.25676.36489
4. Barrera, S.;
Sarango, S.; Montenegro, S. 2019. The phyllosphere microbiome and its potential
application in horticultural crops. A review. Revista Colombiana de Ciencias
Hortícolas. https://revistas.uptc.edu.co/index.php/ciencias_horticolas/article/view/8405
5. Benavides-López,
L. 2019. Cuantificación temprana de Pseudocercospora fijiensis por medio
de qPCR en modelos predictivos de Sigatoka negra en plantas de banano (Musa
AAA). 9-162.
6. Bensch, K.; Braun,
U.; Groenewald, J. Z.; Crous, P. W. 2012. The genus Cladosporium. Studies in
Mycology. 72(1): 1. https://doi.org/10.3114/SIM0003
7. Brown, K.; Hyde,
K.; Guest, D. 1998. Preliminary studies on endophytic fungal communities of Musa
acuminata species complex in Hong Kong and Australia. https://www. fungaldiversity. org/fdp/sfdp/FD_1_27-51.pdf
8. Carrión Abad, B.
A. C. 2015. Análisis microbiológico del complejo orgánico activfol para
determinar la efectividad en el control del hongo (Mycosphaerella fijiensis)
causante de la Sigatoka negra en el banano.
9. Carrion Ramon,
J. J. C. 2020. Monitoreo y control especial de la Sigatoka negra (Mycosphaerella
Fijiensis M), en el cultivo de banano. UTMACH. Unidad Académica de Ciencias
Agropecuarias. Machala. Ecuador.
http://repositorio.utmachala.edu.ec/handle/48000/16135
10. Casas, J.;
López, M.; Salinas, M.; Gisbert, J.; Giménez, E.; García, F.; Sánchez, S.;
Lacalle, A.; Cortés, A.; Moyano, F. 2017. Guía para la realización de un
estudio ambiental: El caso de la cuenca del río Adra (J. Casas, Ed.; Vol. 11).
Universidad de Almería. 3qk3vTP
11. Ceballos, I.;
Mosquera, S.; Angulo, M.; Mira, J.; Argel, L.; Uribe-Velez, D.; Romero-Tabarez,
M.; Orduz-Peralta, S.; Villegas, V. 2012. Cultivable bacteria populations
associated with leaves of banana and plantain plants and their antagonistic
activity against Mycosphaerella fijiensis. Microbial Ecology. 64(3):
641-653. https://doi.org/10.1007/s00248-012-0052-8
12. Cruz-Martín, M.; Acosta-Suárez, M.; Roque, B.; Pichardo, T.;
Castro, R. 2016. Diversidad de cepas bacterianas de la filosfera de Musa spp. con actividad antifúngica frente a Mycosphaerella
fijiensis Morelet. Biotecnología Vegetal. 16(1): 53-60.
13. Cruz-Martín,
M.; Alvarado, Y.; Mena, E.; Acosta, M.; Roque, B.; Pichardo, T. 2018. Cepas
bacterianas con potencial para el manejo de la Sigatoka negra. Anales de la
Academia de Ciencias de Cuba. 8(1).
http://www.revistaccuba.sld.cu/index.php/revacc/article/view/374/373
14. Dissanayake, L.
S.; Wijayawardene, N. N.; Dayarathne, M. C.; Samarakoon, M. C.; Dai, D. Q.;
Hyde, K. D.; Kang, J. C. 2021. Paraeutypella guizhouensis gen. et sp. nov. and Diatrypella
longiasca sp. nov. (Diatrypaceae) from China. Biodiversity Data Journal 9:
e63864. https:// doi.org/10.3897/BDJ.9.E63864
15. Doyle, J. J.;
Doyle, J. L. 1987. A rapid DNA isolation procedure for samal quantities of
fresh leaf tissue. Phytochemical Bulletin. 19: 11-15.
16. Ecuador, M. de
C. E. del. 2017. Informe Sector Bananero Ecuatoriano. Ministerio de Comercio
Exterior. 53(9): 1689-1699. https://doi.org/10.1017/CBO9781107415324.004
17. Etebu, E.;
Young-Harry, W. 2011. Control of black Sigatoka disease: Challenges and
prospects. African Journal of Agricultural Research. 6(3): 508-514.
https://doi.org/10.5897/ AJAR10.223
18. Faleiro, F. G.;
Santos, I. S.; Bahía, R.; Santos, R. F.; Anhert, D. 2002. Otimização da
extração e amplificação de DNA de Theobroma cacao L. visando obtenção de
marcadores RAPD. 31- 34.
19. Fravel, D.;
Olivain, C.; Alabouvette, C. 2003. Fusarium oxysporum and its
biocontrol. New Phytologist. 157(3): 493-502.
https://doi.org/10.1046/J.1469-8137.2003.00700.X
20. García, F.;
Pachacama, S.; Jarrín, D.; Iza, M. 2020. Guía andina para el diagnóstico de
Fusarium Raza 4 Tropical (R4T) Fusarium oxysporum f.sp. Cubense (syn. Fusarium
odoratissimum) agente causal de la marchitez por Fusarium en
musáceas (plátanos y bananos). www.
comunidadandina.org
21. Gauhl, F. 1994.
Epidemiology and ecology of Black Sigatoka on plantain and banana (Musa spp.)
in Costa Rica, Centro América. Ph.D. Thesis of the Systematisch
Geobotanische-Institut der Georg-August-Universität Göttingen and Institut für
Pflanzenpathologie und Pflanzenchutz der Georg-August-Universität Göttingen.
INIBAP.
22. Guzmán, M.
2012. Control biológico y cultural de la sigatoka-negra. Tropical Plant
Pathology. 37 (Suplemento). Agosto. https://doi.org/10.13140/2.1.2927.7442
23. Hao, Y.;
Aluthmuhandiram, J. V. S.; Chethana, K. W. T.; Manawasinghe, I. S.; Li, X.;
Liu, M.; Hyde, K. D.; Phillips, A. J. L.; Zhang, W. 2020. Nigrospora species
associated with various hosts from Shandong Peninsula, China. Mycobiology.
48(3): 169-183. https://doi. org/10.1080/1229 8093.2020.1761747
24.
Hernández-Montiel, L. G.; Larralde-Corona, C. P.; Vero, S.; López-Aburto, M.
G.; Ochoa, J. L.; Ascencio-Valle, F. 2010. Caracterización de levaduras Debaryomyces
hansenii para el control biológico de la podredumbre azul del limón
mexicano. CYTA-Journal of Food. 8(1): 49-56.
https://doi.org/10.1080/19476330903080592
25. Horra, M. L. P.
2014. Evaluación in vitro de la capacidad de extractos orgánicos de
biodiversidad ecuatoriana para inhibir al patógeno Mycosphaerella fijiensis (Morelet)
causante de la Sigatoka Negra en banano.
http://repositorio.puce.edu.ec/bitstream/ handle/22000/10936/4.42.000892.pdf?sequence=4
26. Intriago
Mendoza, L. 2010. Identificación y evolución en laboratorio e invernadero de
microorganismos antagonistas de Sigatoka negra (Mycosphaerella fijiensis
Morelet) en musáceas en el Litoral Ecuatoriano. Tesis de Grado. Universidad
Técnica de Manabí. Ecuador. 95 p.
27. Israeli, Y.;
Lahav, E. 2016. Banana. In Encyclopedia of Applied Plant Sciences. Elsevier. 3:
363-381. https://doi.org/10.1016/B978-0-12-394807-6.00072-1
28. Izzeddin, A.
N.; Medina, T. L. 2011. Efecto del control biológico por antagonistas sobre
fitopatógenos en vegetales de consumo humano. Salus. 15(3): 8-18.
29. Lima, N. B.;
Marcus, M. V.; Morais, M. A. D.; Barbosa, M. A. G.; Michereff, S. J.; Hyde, K.
D.; Câmara, M. P. S. 2013. Five Colletotrichum species are responsible for
mango anthracnose in northeastern Brazil. Fungal Diversity. 61(1): 75-88.
https://doi.org/10.1007/s13225- 013-0237-6
30. López, S. 2019.
Bacillus un género que alberga especies que cumplen diversos roles biológicos.
Infive-Conicet UNLP. 1-26.
31. Martínez, G.;
Pargas, R.; Manzanilla, E. 2012. Orden Zingiberales: Las musáceas y su relación
con plantas afines. Scielo.
http://ve.scielo.org/scielo.php?script=sci_arttext&pid=S0002-
192X2012000100014
32. Morillo, E.;
Miño, G. 2011. Marcadores moleculares en biotecnología agrícola: manual de
técnicas y procedimientos en INIAP.
https://repositorio.iniap.gob.ec/bitstream/41000/848/4/ iniapscm91.pdf
33. Moubasher, A.
H.; Moharram, A. M.; Ismail, M. A.; Abdel-Hafeez, M. H. 2016. Aeromycobiota
over banana plantations in Assiut Governorate and enzymatic producing potential
of the most common species. Journal of Basic & Applied Mycology (Egypt). 7:
9-18.
34. Muimba-Kankolongo, A. 2018. Fruit Production. In Food crop
production by smallholder farmers in Southern Africa (p. 275-312). Elsevier.
https://doi.org/10.1016/B978-0-12-814383- 4.00012-8
35. Nam, M. H.;
Park, M. S.; Kim, H. S.; Kim, T. I.; Kim, H. G. 2015. Cladosporium
cladosporioides and C. tenuissimum cause blossom blight in
strawberry in Korea. Mycobiology. 43(3): 354.
https://doi.org/10.5941/MYCO.2015.43.3.354
36. Novák, P.;
Hřibová, E.; Neumann, P.; Koblížková, A.; Doležel, J.; Macas, J. 2014.
Genome-wide analysis of repeat diversity across the family musaceae. PLoS ONE.
9(6). https://doi.org/10.1371/ journal.pone.0098918
37. Nuangmek, W.;
McKenzie, E. H. C.; Lumyong, S. 2008. Endophytic fungi from wild banana (Musa
acuminata Colla) works against anthracnose disease caused by Colletotrichum
musae. Research Journal of Microbiology. 3(5): 368-374. https://doi.
org/10.3923/ JM.2008.368.374
38. Photita, W.;
Lumyong, S.; Lumyong, P.; Mckenzie, E. H. C.; Hyde, K. D.; Photita, W.;
Lumyong, S.; Lumyong, P.; Hyde, M. E. H. C. 2004. Fungal diversity are some
endophytes of Musa acuminata latent pathogens? Fungal Diversity. 16:
131-140.
39. Pinheiro da
Costa, S.; Schuenck-Rodrigues, R.; da Silva Cardoso, V.; Valverde, S.;
Vermelho, A.; Ricci-Júnior, E. 2021. Actividad antimicrobiana de hongos
endofíticos aislados de Brugmansia suaveolens Bercht.; Presl, J.
Research, Society and Development. 10(14): e113101421646. https://doi.
org/10.33448/rsd-v10i14.21646
40. Pitt, W. M.;
Trouillas, F. P.; Gubler, W. D.; Savocchia, S.; Sosnowski, M. R. 2013.
Pathogenicity of Diatrypaceous Fungi on grapevines in Australia. Plant disease.
97(6): 749-756. https:// doi.org/10.1094/PDIS-10-12-0954-RE
41. Poveda, I.;
Cruz-Martín, M.; Sánchez-García, C.; Acosta-Suárez, M.; Leiva-Mora, M.; Roque,
B.; Alvarado-Capó, Y. 2010. Caracterización de cepas bacterianas aisladas de la
filosfera de Musa spp. con actividad antifúngica in
vitro frente a Mycosphaerella fijiensis. Biotecnología Vegetal.
10(1): 57-61.
42. Ramírez, M.;
Rodriguez, T.; Peniche, C.; Alfonso, L. 2010. Chitin and its derivatives as
biopolymers with potential agricultural applications. Biotecnología Aplicada.
27: 270-276.
43. Riera, N. J.
2015. Caracterización molecular y de patogenicidad de Colletotrichum spp,
en bananas var Cavendish y pruebas de antagonismo con Trichoderma spp.,
recolectadas en fincas bananeras de la región costa del Ecuador. Universidad
San Francisco de Quito. 98.
44. Ruiz-Boyer, A.;
Rodríguez-González, A. 2020. Preliminary list of fungi (Ascomycota and
basidiomycota) and myxomycetes (myxomycota) of Isla del Coco, Puntarenas, Costa
Rica. Revista de Biología Tropical. 68(S1): S33-S56.
https://doi.org/10.15517/RBT. V68IS1.41165
45. SIPA. 2021.
Ficha del cultivo de banano (Musa paradisiaca AAA).
https://sipa.agricultura.gob.ec/ index.php/bananos
46. Šišić, A.;
Baćanović-Šišić, J.; Al-Hatmi, A. M. S.; Karlovsky, P.; Ahmed, S. A.; Maier,
W.; Hoog, G. S. D.; Finckh, M. R. 2018. The ‘forma specialis’ issue in Fusarium:
A case study in Fusarium solani f. Sp. Pisi. Scientific Reports 2018
8:1, 8(1), 1-17. https://doi.org/10.1038/s41598-018- 19779-z
47. Sun, X.; Guo,
L. D. 2012. Endophytic fungal diversity: Review of traditional and molecular
techniques. Mycology. 3(1): 65-76. https://doi.org/10.1080/21501203.2012.656724
48. Suwannarach,
N.; Kumla, J.; Bussaban, B.; Nuangmek, W.; Matsui, K.; Lumyong, S. 2013.
Biofumigation with the endophytic fungus Nodulisporium spp. CMU-UPE34 to
control postharvest decay of citrus fruit. Crop Protection. 45: 63-70. https://doi.org/10.1016/J. CROPRO.2012.11.015
49. Toju, H.;
Tanabe, A. S.; Yamamoto, S.; Sato, H. 2012. High-Coverage ITS Primers for the
DNA-Based Identification of Ascomycetes and Basidiomycetes in Environmental
Samples. PLoS ONE. 7(7): 40863. https://doi.org/10.1371/JOURNAL.PONE.0040863
50. Trouillas, F.
P.; Úrbez-Torres, J. R.; Gubler, W. D. 2017. Diversity of diatrypaceous fungi
associated with grapevine canker diseases in California. 102(2): 319-336.
https://doi.org/10.3852/08- 185
51. Vázquez, J.;
González, J.; Castillo, J.; Álvarez, M. 2019. Microorganismos benéficos MOBs
obtenidos de plantas, como promotores en la germinación de semillas. Dominio de
Las Ciencias. 5: 615-628. http://dx.doi.org/10.23857/dc.v5i1.1064 Cie
52. Velasco, R.;
Tapia, R. 2014. Curso práctico de Microbiología. Universidad Autónoma
Metropolitana.
53. Vilgalys, R.
2003. Taxonomic misidentification in public DNA databases. New Phytologist.
160(1): 4-5. https://doi.org/10.1046/J.1469-8137.2003.00894.X
54. Villamil
Carvajal, J. E. V.; Viteri Rosero, S. E. V.; Villegas Orozco, W. L. V. 2015.
Aplicación de antagonistas microbianos para el control biológico de Moniliophthora
roreri Cif & Par en Theobroma cacao L. bajo condiciones de
campo. Revista Facultad Nacional de Agronomía Medellín. 68(1): 7441-7450.
https://doi.org/10.15446/rfnam.v68n1.47830
55.
Villegas-Escobar, V.; Ceballos, I.; Mira, J. J.; Argel, L. E.; Peralta, S. O.;
Romero-Tabarez, M. 2013. Fengycin C produced by Bacillus subtilis EA-CB0015.
Journal of Natural Products. 76(4): 503-509. https://doi.org/10.1021/np300574v
56. Walker, C.; Muniz, M. F. B.; Rolim, J. M.; Martins, R. R.
O.; Rosenthal, V. C.; Maciel, C. G.; Mezzomo, R.; Reiniger, L. R. S. 2016.
Morphological and molecular characterization of Cladosporium cladosporioides
species complex causing pecan tree leaf spot. Genetics and molecular
research: GMR. 15(3). https://doi.org/10.4238/GMR.15038714
57. Wang, M.; Liu,
F.; Crous, P. W.; Cai, L. 2017. Phylogenetic reassessment of Nigrospora:
Ubiquitous endophytes, plant and human pathogens. Persoonia: Molecular
Phylogeny and Evolution of Fungi. 39(December). 118-142.
https://doi.org/10.3767/persoonia.2017.39.06
58. Zakaria, L.; Aziz, W. N. W. 2018. Molecular Identification
of endophytic fungi from banana leaves (Musa spp.). Tropical Life Sciences
Research. 29(2): 201. https://doi.org/10.21315/ TLSR2018.29.2.14