Revista de la Facultad de Ciencias
Agrarias. Universidad Nacional de Cuyo. Tomo 57(1). ISSN (en línea) 1853-8665.
Año 2025.
Original article
Arachis
genetic resources: evaluation of peanut smut resistance in wild
species
Recursos
genéticos de Arachis: evaluación de resistencia al carbón de maní en
especies silvestres
Melina Hebelen
Rosso1,
Damián Francisco
Giordano2,
Claudio Oddino3,
Alejandra V. García4,
Sara Soave1,
Juan Soave1,
1 Criadero El Carmen. Av. Italia 871. C. P. 5809. General
Cabrera. Córdoba. Argentina.
2 IMICO, CONICET- UNRC. RN 36 Km 601. C. P. 5804. Río Cuarto.
Córdoba. Argentina.
3 Universidad Nacional de Río Cuarto. Facultad de Agronomía y
Veterinaria. RN 36 Km 601. C. P. 5804. Río Cuarto. Córdoba. Argentina.
4 IBONE, CONICET - UNNE. Sargento Juan Bautista Cabral 2131. C.
P. 3400. Corrientes. Argentina.
5 FaCENA. UNNE. Av. Libertad 5460. C. P. 3400. Corrientes.
Argentina.
* graciela.lavia@yahoo.com.ar
Abstract
Genetic resources
are essential for crop improvement. Particularly, wild species related to
peanuts are an important source of resistance to various factors. Thecaphora
frezii, a pathogen causing peanut smut, leads to yield losses in
Argentina’s peanut sector up to 35%. This study evaluated the response of 11
diploid species with A, B, F and K genomes, A. monticola (AABB), and
diploid interspecific hybrids (BB), to T. frezii over two cropping
seasons. Plants were grown in 20L pots (three replicates each) under field
conditions and inoculated with teliospores of the pathogen (20,000 tel./g of
soil). The disease was quantified through incidence (% of diseased pods) and
severity (scale from 0 to 4). Among A genome species, A. duranensis exhibited
the highest incidence at 15.27%; for K genome species, A. batizocoi reached
13.18%. Resistance to T. frezii was observed in the wild species A.
diogoi and A. stenosperma (A genome), A. williamsii (B
genome), A. trinitensis (F genome), A. cruziana (K genome), and
the intragenomic hybrids, constituting new records. Our findings expand the
peanut gene pool information for breeders and identify resistant genotypes,
supporting the need to preserve wild peanut germplasm to ensure its
availability.
Keywords: Thecaphora frezii, Arachis
hypogaea, wild peanut, resistance, genomes
Resumen
Los recursos
genéticos son fundamentales para el mejoramiento de los cultivos.
Particularmente, las especies silvestres afines al maní cultivado constituyen
una valiosa fuente de resistencias. Thecaphora frezii Carranza &
Lindquist ocasiona pérdidas en el sector manisero argentino de aproximadamente
el 35% del rendimiento. Se evaluó, durante dos campañas, el comportamiento
frente a T. frezii de 11 especies diploides con genomas AA, BB, FF y KK;
A. monticola AABB, y un híbrido interespecífico diploide BB. Los materiales
se sembraron en macetas de 20L (tres repeticiones por c/u), en condiciones de
campo, inoculándose con teliosporas del patógeno (20000 tel./g de suelo). Se
cuantificó la enfermedad mediante la incidencia (% de vainas enfermas) y la
severidad (escala de 0 a 4). Entre las especies con genoma A, A. duranensis presentó
la mayor incidencia, 15,27%; y en las de genoma K, A. batizocoi, 13,18%.
La resistencia a T. frezii hallada en las especies silvestres A.
diogoi y A. stenosperma (genoma A), A. williamsii (genoma B),
A. trinitensis (genoma F), A. cruziana (genoma K) y en el híbrido
intragenómico BB constituyen nuevos registros. Nuestros resultados permiten
ampliar el acervo genético del maní y generar genotipos resistentes;
ratificando que el germoplasma de maní silvestre debe preservarse
cuidadosamente para asegurar su disponibilidad.
Palabras clave: Thecaphora frezii, Arachis
hypogaea, maníes silvestres, resistencia, genomas
Originales: Recepción: 28/08/2024 - Aceptación: 27/11/2024
Introduction
Peanut (A.
hypogaea L.) is an allotetraploid species (2n=4x=40, AABB) originated in
South America. It is cultivated in warm regions worldwide, with an annual
production of 45.5 million tons (32). Given the small
intern market, Argentina exports approximately 80% of its production. Córdoba
province accounts for nearly 90% of Argentina’s peanut industry (28).
The fungus Thecaphora
frezii Carranza & Lindquist causes peanut smut, an endemic disease in
Argentina (23) first detected in
commercial crops in north-central Córdoba (18). Symptoms include
pod malformation and replacement of seeds by dark-brown teliospores (figure 1). Literature
reports almost 50% incidence (21) and yield losses
up to 35% (20, 23).

Powder inside the
pods evidences Thecaphora frezii teliospores. Scale bar = 1 cm.
El
polvillo que se observa en el interior son las teliosporas de Thecaphora
frezii. Escala de barra = 1 cm.
Figure
1. Arachis duranensis pods
(K 7988) with smut (Severity Scale: 4).
Figura
1. Vainas de A. duranensis (K
7988) afectadas con carbón (Grado de Severidad: 4).
Current management
practices, such as tillage, crop rotation, cultivar selection, fungicide and
fertilizer applications or soil amendments, have had limited success in
reducing yield losses (1, 11, 22). However, the
development of recombinant inbred lines (RILs) has enabled breeding strategies,
generating resistant genotypes (8). These RILs
originated from crosses involving one synthetic amphidiploid parent, obtained
from a triple cross among wild species [(A. correntina x A.
cardenasii) x A. batizocoi)]4x,
and an experimental line of A. hypogaea. These findings highlight the
importance of wild species as biotic resistance sources in breeding strategies,
targeting novel genetic resources with stable resistance to peanut smut.
Wild Arachis diploid
species with A, B, D, F y K genomes (24, 25, 29, 30), are
phylogenetically close to cultivated peanuts, constituting the genetic
secondary pool PG-2 (14), defined as those
species that can be crossed with local cultivars/elite germplasm, to produce
fertile F1. Some of these species have been previously evaluated for smut
resistance (3). Most of them are
preserved in the “Banco de Germoplasma BGCTES” (IBONE, FCA-UNNE) germplasm
bank. Although these genetic resources constitute fundamental breeding
resources (31), they have not yet
been fully exploited.
This study evaluated wild Arachis species with A, B, F y
K genomes (24, 25)
and intragenomic reciprocal hybrids (10),
conserved in the BGCTES germplasm bank, aiming to identify wild sources of
resistance against peanut smut.
Materials
and Methods
Eleven diploid species with AA, BB, FF, and KK genomes, a
tetraploid A. monticola AABB, and two intragenomic diploid hybrids (table
1) were evaluated over two cropping seasons (2019/2020 and
2020/2021), under field conditions at Criadero El Carmen in General Cabrera,
Córdoba, Argentina (32°49’39.49” S - 63°51’55.57” O).
Table 1. Analyzed
material (species/hybrids, collection data and genome) and disease evaluation
results.
Tabla 1. Material
analizado (especies/híbridos, datos de colección y genoma) y resultados
obtenidos de la evaluación de la enfermedad.
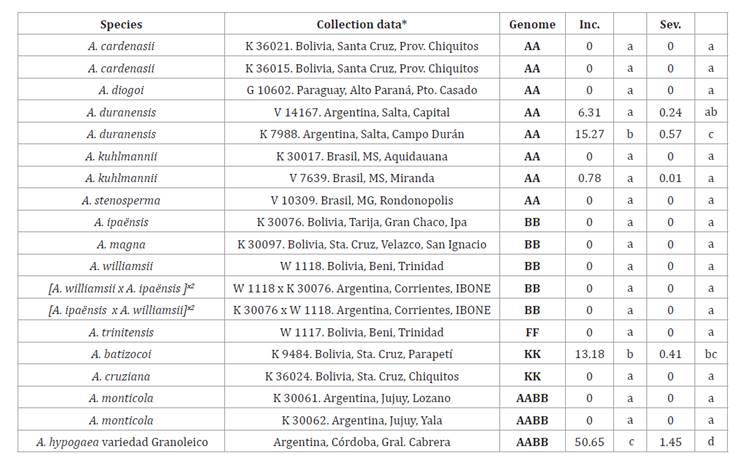
Except for the control, the
analyzed materials are stored at the BGCTES (Corrientes, Argentina).
* G, W.C. Gregory; K, A.
Krapovickas; V, J.F.M Valls; W, D.E. Williams; Inc. incidence (%), Sev.
Severity (0-4). Different letters in each column indicate statistically
significant differences (p ≤ 0.05).
Los materiales analizados se
mantienen en el BGCTES (Corrientes, Argentina), excepto el control.
* G, W.C.
Gregory; K, A. Krapovickas; V, J.F.M Valls; W, D.E. Williams. ; V, J.F.M Valls;
W, D.E. Williams. Inc. Incidencia (%), Sev. Severidad (0-4). Letras diferentes
indican diferencias estadísticamente significativas (p ≤ 0,05).
The species were sown in 20L pots during the first week of
December and maintained under field conditions in a completely randomized
design with three replicates. Susceptible A. hypogaea var. Granoleico
was included as a control treatment. Teliospores obtained from infected pods
were used to inoculate the pots at a concentration of 20000tel/g soil. During
harvest in April, pods were manually opened to quantify disease incidence
(percentage of infected pods) and severity, following the 0 to 4 scale proposed
by Astiz Gassó et al. (2008),
where 0=healthy pod, 1 = normal pod with initial seed infection, 2 = normal pod
with 50% seed infection, 3 = normal pod with 75% seed infection, and 4 =
deformed pod with 100% seed infection. Severity was calculated using equation
1.
where
(x0-x4)
= the number of fruits in each classification
(0-4) = the
classification number.
Data were analyzed using ANAVA and Duncan´s test (p≤0.05) with
InfoStat software 2020 (9).
Results
Only species with AA and KK genomes exhibited symptoms of fungal
infection. Among AA species, both entries of A. duranensis and one of A.
kuhlmannii showed affected pods, with A. duranensis K 7988
presenting the highest average incidence of 15.27% (figure 1).
Among KK genome species, A. batizocoi showed an average incidence of
13.18%, while A. cruziana showed no affected pods. Conversely, no
affected pods were observed in species with BB and FF genomes, in the
tetraploid A. monticola AABB, or BB hybrids [A. ipaënsis x A.
williamsii]2x and its reciprocal cross.
The control, A. hypogaea var. Granoleico, displayed an average incidence
of 50.65%. This value, along with those obtained for the wild species A.
duranensis and A. batizocoi, showed statistically significant
differences with the other wild species and the interspecific hybrid results (table
1).
Regarding severity
analysis in wild species, A. duranensis K 7988 achieved the highest
value at 0.57, followed by A. batizocoi, with an average of 0.41. The
control species, A. hypogaea var. Granoleico, had an incidence value of
1.45. Statistical analysis revealed significant differences (p≤0.05) between
these values and those obtained for the rest of the species evaluated.
Discussion
The Arachis genus
includes nine infrageneric sections according to cross-compatibility and
exomorphic traits (17). Among these, the
Arachis section is notable for comprising the largest number of wild species
(32 spp.), and for its economic importance, as it includes the cultivated
peanut A. hypogaea. In this section, 15 species possess A genome, six
have B genome, K and G genomes are represented by three species each, F genome
is represented by 2 species and only one has D genome (24,
25, 26, 29, 30). According to Harlan and Wet (1971), all 32 species
integrate A. hypogaea secondary gene pool (PG-2).
Wild diploid Arachis species constitute valuable
gene-transfer resources for cultivated peanuts, providing resistance to biotic
and abiotic factors. Several techniques and methodologies have been developed
for gene introgression from wild to cultivated genotypes (7,
31). In Argentina, the introgression of resistance to peanut smut
from wild species has allowed important breeding advancements, such as the
development of EC - 191 RC (AO) and EC - 394 RC (AO); (19).
Our
results revealed a close relation between genome types and resistance to T.
frezii. Accessions with A genome responded as previously observed by De
Blas et al. (2019), except A. duranensis.
Previous evaluations indicated susceptibility in A. duranensis (3),
which aligns with our results, thus constituting the first susceptible A genome
species identified. In our tests involving K genome species, A. cruziana was
non-susceptible, while A. batizocoi exhibited susceptibility, which
contrasts with prior results (8).
Despite that, values were not markedly higher than those reported in this work.
Species with B genome, alongside with [A. ipaënsis x A. williamsii]x2 and the reciprocal
hybrid, showed non-infected pods, suggesting that B genome would be resistant
to T. frezii, as previously reported (8).
Resistance
to Thecaphora frezii in the wild species A. diogoi, A.
stenosperma (A genome), A. williamsii (B genome), A. trinitensis (F
genome), A. cruziana (K genome), and the diploid intragenomic hybrids
constitute new records for Arachis genus.
The
identification of resistant resources offers new breeding opportunities for
peanut improvement. The literature documents successful incorporations of wild Arachis
species in breeding programs. For instance, the A genome species A.
cardenasii and A. stenosperma, which exhibit resistance to T.
frezii, have also shown resistance to nematodes, rust and leaf spots (31).
NemaTAM, a nematode-resistant genotype, was developed from wild A.
cardenasii (27).
Additionally, resistance to Meloidogyne arenaria was successfully
transferred from A. stenosperma to tetraploid peanut (4).
Considering
tetraploids, A. hypogaea is a segmentary allotetraploid with 2n=40
chromosomes (6, 13, 15, 16).
The AABB cultigen arose through interspecific hybridization of two diploid AA
and BB species (A. duranensis and A. ipaënsis, respectively),
followed by chromosomal duplication (5, 6, 12).
This event originated the wild tetraploid ancestor A. monticola, which
subsequently underwent domestication, resulting in the cultigen A. hypogaea.
In this study, our control species A. hypogaea exhibited the highest
value of disease incidence and severity, whereas A. monticola showed no
susceptibility. Given the susceptibility of A. duranensis to T.
frezii, we hypothesize that A. monticola´s resistance may be derived
from the B genome in A. ipaënsis, a hypothesis that could be further
explored.
Conclusions
Our results provide valuable insights into wild Arachis species
as sources of resistance to peanut smut disease, enabling breeders to expand
peanut genetic pool and develop resistant genotypes. This underscores the
importance of carefully preserving wild peanut germplasm collections to ensure
their availability for future breeding efforts.
Acknowledgements
This work was supported by the Agencia Nacional de Promoción
Científica y Técnica (PICT-2015-2681) and the Secretaría General de Ciencia y
Técnica de la Universidad Nacional del Nordeste (PI 6P003).
1. Arias, S. L.;
Mary, V. S.; Velez, P. A.; Rodriguez, M. G.; Otaiza-Gonzalez, S. N.; Theumer,
M. G. 2021. Where does the peanut smut pathogen, Thecaphora frezii, ft
in the spectrum of smut diseases? Plant Dis. 105(9): 2268-2280.
doi.org/10.1094/PDIS-11-20-2438-FE
2. Astiz Gassó, M.;
Leis, R.; Marinelli, A. 2008. Evaluación de incidencia y severidad del carbón
de maní (Thecaphora frezii) en infecciones artificiales, sobre
cultivares comerciales de maní. Actas 1° Congreso Argentino de Fitopatología.
Córdoba. http://aafitopatologos.com.ar/wp/wp-content/uploads/2014/11
3. Astiz Gassó, M.;
Leis, R.; Marinelli, A. 2011. Evaluación de la tolerancia del germoplasma de
maní (Arachis spp.) para el manejo del carbón Thecaphora frezii.
Actas XXVI Jornada Nacional del maní. Argentina.
4. Ballen-Taborda,
C.; Chu, Y.; Ozias-Akins, P.; Timper, P.; Holbrook, C. C.; Jackson, S. A.;
Bertioli, D. J.; Bertioli, S. C. M. 2019. A new source of root-knot nematode
resistance from Arachis stenosperma incorporated into allotetraploid
peanut (Arachis hypogaea). Scientific Reports. 9: 17702.
https://doi.org/10.1038/s41598-019-54183-1
5. Bertioli, D. J.; Cannon, S. B.; Froenicke, L.; Huang, G.; A.
D. Farmer, E. K. S. Cannon; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; Ren, L.;
Moretzsohn, M. C.; Shirasawa, K.; Huang, W.; Vidigal, B.; Abernathy, B.; Chu,
Y.; Niederhuth, C. E.; Umale, P.; Araújo, A. C. G.; Kozik, A.; Do Kim, K.;
Burow, M. D.; Varshney, R. K.; Wang, X.; Zhang, X.; Barkley, N.; Guimarães, P.
M.; Isobe, S.; Guo, B.; Liao, B.; Stalker, H. T.; Schmitz, R. J; Scheffler, B.
E.; Leal-Bertioli, S. C. M.; Xum, X.; Jackson, S. A.; Michelmore, R.;
Ozias-Akins, P. 2016. The genome sequences of Arachis duranensis and Arachis
ipaënsis, the diploid ancestors of cultivated peanut. Nat. Genet. 48:
438-446. doi:10.1038/ng.3517
6. Bertioli, D. J.;
Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli,
S. C. M.; Ren, L.; Farmer, A. D.; Pandey , M. K.; Samoluk, S. S.; Abernathy,
B.; Agarwal, G.; Ballén-Taborda, C.; Cameron, C.; Campbell, J.; Chavarro, C.;
Chitikineni, A.; Chu, Y.; Dash, S.; Baidouri, M. E.; Guo, B.; Huang, W.; Do
Kim, K.; Korani , W.; Lanciano, S.; Lui, C. G.; Mirouze, M. ; Moretzsohn, M.
C.; Pham, M.; Shin, J. H.; Shirasawa, K.; Sinharoy, S.; Sreedasyam , A.; Weeks
, N. T.; Zhang, X.; Zheng, Z.; Sun, Z.; Froenicke, L.; Aiden, E. L.;
Michelmore, R.; Varshney, R. K.; Holbrook, C. C.; Cannon , E. K. S.; Scheffler,
B. E.; Grimwood, J.; Ozias-Akins, P.; Cannon, S. B.; Jackson, S. A.; Schmutz,
J. 2019. The genome sequence of segmental allotetraploid peanut Arachis
hypogaea. Nat. Genet. 51: 877-884.
7. Cason, J. M.;
Simpson, C. E.; Burow, M. D.; Tallury, S.; Pham, H.; Ravelombola, S. W. 2023.
Use of wild and exotic germplasm for resistance in peanut. Journal of Plant
Registrations. 17(1): 1-25. https://doi.org/10.1002/plr2.20261
8. de Blas, F. J.;
Bressano, M.; Teich, I.; Balzarini, M. G.; Arias, R. S.; Manifesto, M. M.;
Costero, B. P.; Oddino, C.; Soave, S. J.; Soave, J. A.; Buteler, M. I.; Massa,
A. N.; Seijo, J. G. 2019. Identification of smut resistance in wild Arachis species
and its introgression into peanut elite lines. Crop Science. 59:1657-1665. doi:
10.2135/cropsci2018.10.0656
9. Di Rienzo, J.
A.; Casanoves, F.; Balzarini, M. G.; González, L.; Tablada, M.; Robledo, C. W.
2020. InfoStat, versión 2020. Centro de Transferencia InfoStat. Universidad
Nacional de Córdoba. Argentina. http://www.infostat.com.ar
10. García, A. V.;
Silvestri, M. C.; Vandecaveye, M. A.; Custodio, A. R.; Moretzsohn, M. C.;
Lavia, G. I. 2021. Genomic affinity in hybrids of B-genome Arachis species:
new genetic resources toward peanut improvement. Crop Breeding and Applied
Biotechnology. 21(3): e38292139. http://dx.doi.org/10.1590/1984-
70332021v21n3a48
11. Giordano, D.
F.; Del Canto, A.; Erazo, J. G.; Pastor, N. A.; Crenna, A. C.; Rosso, M.;
Torres, A. M.; Oddino, C. M. (en prensa). Fungicide management of late leaf
spot and peanut smut. Revista de la Facultad de Ciencias Agrarias. Universidad
Nacional de Cuyo. Mendoza. Argentina.
12. Grabiele, M.;
Chalup, L.; Robledo, R.; Seijo, G. 2012. Genetic and geographic origin of
domesticated peanut as evidenced by 5S rDNA and chloroplast DNA sequences.
Plant Systematics and Evolution. 298: 1151-1165. Doi: 10.1007/s00606-012-0627-3
13. Gregory, W. C.;
Gregory, M. P. 1976. Groundnut. En: Simmonds, N. W. (Ed.). Evolution of crop
plants. London. Longman Group Ltd. 151-154.
14. Harlan, J. R.;
de Wet, J. M. J. 1971. Toward a rational classification of cultivated plants.
Taxon. 20(4): 509-517.
15. Husted, L.
1933. Cytological studies of the peanut Arachis I. Chromosome number,
and morphology. Cytologia. 5: 109-117. https://doi.org/10.1508/cytologia.5.109
16. Husted, L.
1936. Cytological studies of the peanut Arachis II. Chromosome number,
morphology and behavior and their application to the problem of the origin of
cultivated forms. Cytologia. 7: 396-423. ttps://doi.org/10.1508/cytologia.7.396
17. Krapovickas,
A.; Gregory, W. C 1994. Taxonomía del género Arachis (Leguminosae).
Bonplandia. 8: 1-186.
18. Marinelli, A.;
March, G.; Rago, A. M. 1995. El carbón del maní Thecaphora frezii sobre Arachis
hypogaea. Actas VII Congreso de Micología y XVII Jornadas Argentinas de
Micología. Argentina.
19. Oddino, C.;
Rosso, M.; Giordano, D. F.; de Blas, F.; Bressano, M.; Soave, S.; Moresi, A.;
Seijo, G.; Buteler; M.; Soave, J. 2021. Comportamiento a enfermedades y
rendimiento de genotipos provenientes de maníes silvestres. Actas XXXVI Jornada
Nacional del Maní. Argentina.
20. Paredes, J. A.;
Cazón, L. I.; Osella, A.; Peralta, V.; Alcalde, M.; Kearney, M. I.; Zuza, M.
S.; Rago, A. M.; Oddino, C. 2016. Relevamiento regional del carbón de maní y
estimación de pérdidas producidas por la enfermedad. Actas XXXI Jornada
Nacional de Maní. Argentina.
21. Paredes, J. A.;
Guzzo, M. C.; Monguillot, J. H.; Asinari, F.; Posada, G. A.; Oddino, C. M.;
Giordano, D. F.; Morichetti, S. A.; Torres, A. M.; Rago, A. M.; Monteoliva, M.
I. 2024. Low water availability increases susceptibility to peanut smut (Thecaphora
frezzii) in peanut crop. Plant Pathology. 73: 316-325.
https://doi.org/10.1111/ppa.13810
22. Pedelini, R.
2016. Maní, guía práctica para su cultivo. Boletín de divulgación técnica N° 2.
4° Edición.
23. Rago, A. M.;
Cazón, L. I.; Paredes, J. A.; Edwards Molina, J. P.; Conforto, E. C.; Bisonard,
E. M.; Oddino, C. 2017. Peanut smut: from an emerging disease to an actual
threat to Argentine peanut production. Plant Disease. 101: 400-408.
24. Robledo, G.;
Lavia, G. I.; Seijo, J. G. 2009. Species relations among wild Arachis species
with the A genome as revealed by FISH mapping of rDNA loci and heterochromatin
detection. Theoretical and Applied Genetics. 118: 1295-1307.
doi:10.1007/s00122- 009-0981-x
25. Robledo, G.;
Seijo, J. G. 2010. Species relationships among the wild B genome of Arachis species
(section Arachis) based on FISH mapping of rDNA loci and heterochromatin
detection: A new proposal for genome arrangement. Theoretical and Applied
Genetics. 121: 1033-1046. doi:10.1007/s00122-010-1369-7
26. Silvestri, M.
C.; Ortiz, A. M.; Lavia, G. I. 2015. rDNA loci and heterochromatin positions
support a distinct genome type for ‘x = 9 species of section Arachis (Arachis,
Leguminosae). Plant Systematics and Evolution. 301: 555-562.
doi:10.1007/s00606-014-1092-y
27. Simpson, C. E.; Starr J. L.; Church, G. T.; Burrow, M. D.;
Paterson, A. H. 2003. Registration of NemaTAM peanut. Crop Science. 43: 1561.
doi:10.2135/cropsci2003.1561
28. SISA. 2023.
Sistema de Información Simplificado Agrícola. https://www.afip.gob.ar/
actividadesAgropecuarias/sector-agro/sisa/informacion-productiva.asp
29. Smartt, J.;
Gregory, W. C.; Gregory, M. P. 1978. The genomes of Arachis hypogaea. 1.
Cytogenetic studies of putative genome donors. Euphytica. 27: 665-675.
https://doi.org/10.1007/ BF00023701
30. Stalker, H. T.
1991. A new species in section Arachis of peanut with a D genome.
American Journal of Botany. 78: 630-637.
https://doi.org/10.1002/j.1537-2197.1991.tb12587.x
31. Stalker, H. T.
2017. Utilizing wild species for peanut improvement. Crop Science. 57:
1102-1120. doi:10.2135/cropsci2016.09.0824
32. USDA. 2023. United States Department of Agriculture. Peanut
explorer. https://ipad.fas. usda.gov/cropexplorer/cropview/commodityView.aspx?cropid=2221000&sel_
year=2022&rankby=Production. Consultado 12/09/2023.